C-930 lot 211005
RSP 12602
Grower: NSI lab solutions
General Information

- Sample Name
- NSI Hemp
- Accession Date
- February 10, 2022
- Reported Plant Sex
- not reported
- Report Type
- CannSNP90
The strain rarity visualization shows how distant the strain is from the other cultivars in the Kannapedia database. The y-axis represents genetic distance, getting farther as you go up. The width of the visualization at any position along the y-axis shows how many strains there are in the database at that genetic distance. So, a common strain will have a more bottom-heavy shape, while uncommon and rare cultivars will have a visualization that is generally shifted towards the top.
Chemical Information
Cannabinoid and terpenoid information provided by the grower.
Cannabinoids
No information provided.
Terpenoids
No information provided.
Genetic Information
- Plant Type
- Type III
File Downloads
This chart represents the Log-R Ratio (LRR) over variants in the region of the THCA synthase gene. A high correlation between the LRR and samples with a known deletion in THCA synthase indicates the THCA region is deleted and a low correlation indicates it is intact.
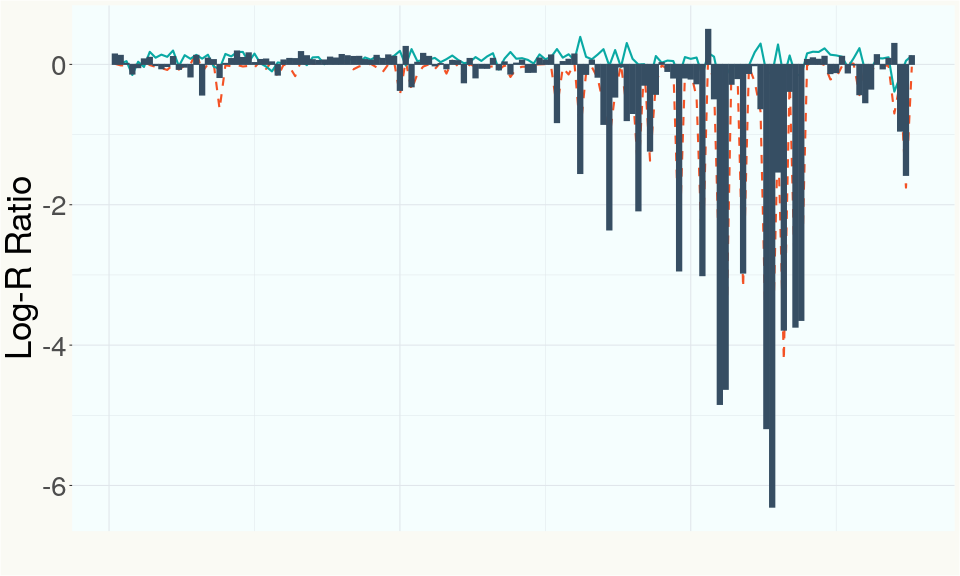
This chart represents the Log-R Ratio (LRR) over variants in the region of the CBDA synthase gene. A high correlation between the LRR and samples with a known deletion in CBDA synthase indicates the CBDA region is deleted and a low correlation indicates it is intact.
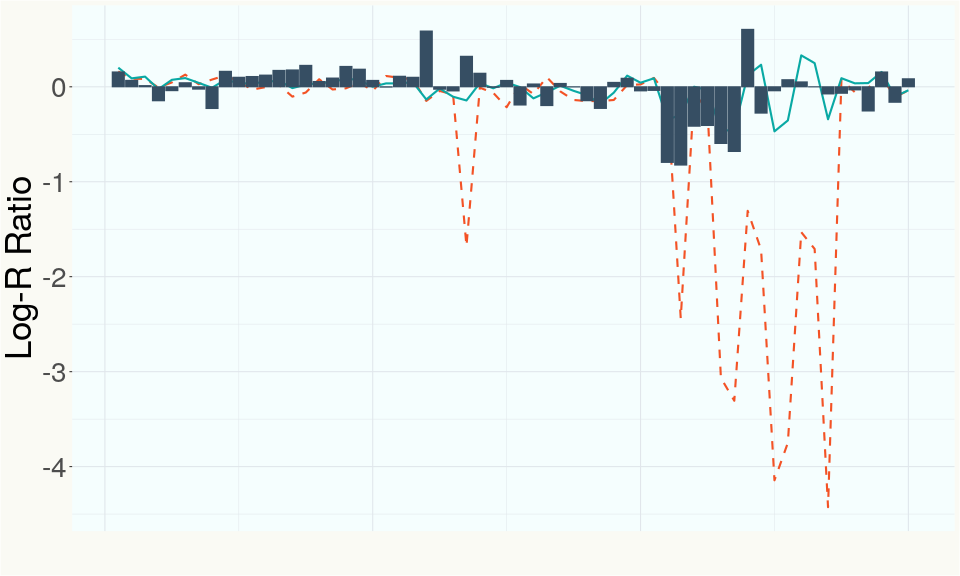
This chart represents the Log-R Ratio (LRR) over variants in the region of the CBCA synthase gene. A high correlation between the LRR and samples with a known deletion in CBCA synthase indicates the CBCA region is deleted and a low correlation indicates it is intact.
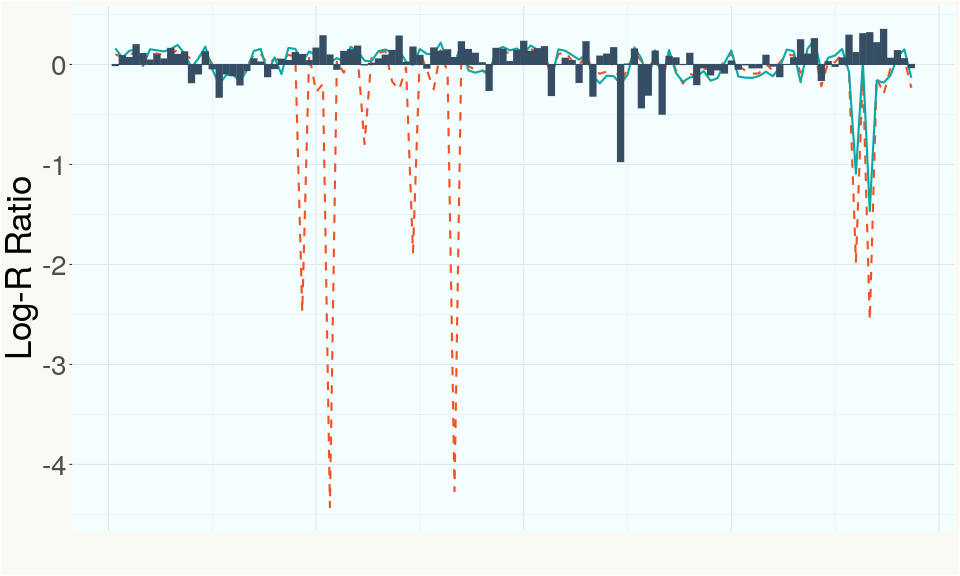
This chart represents the Log-R Ratio (LRR) over variants in the Y-contigs. A high correlation between the LRR and samples which are known Females indicates these Y-contigs are deleted in this sample and a low correlation indicates that the Y-contigs are not deleted and is likely Male.
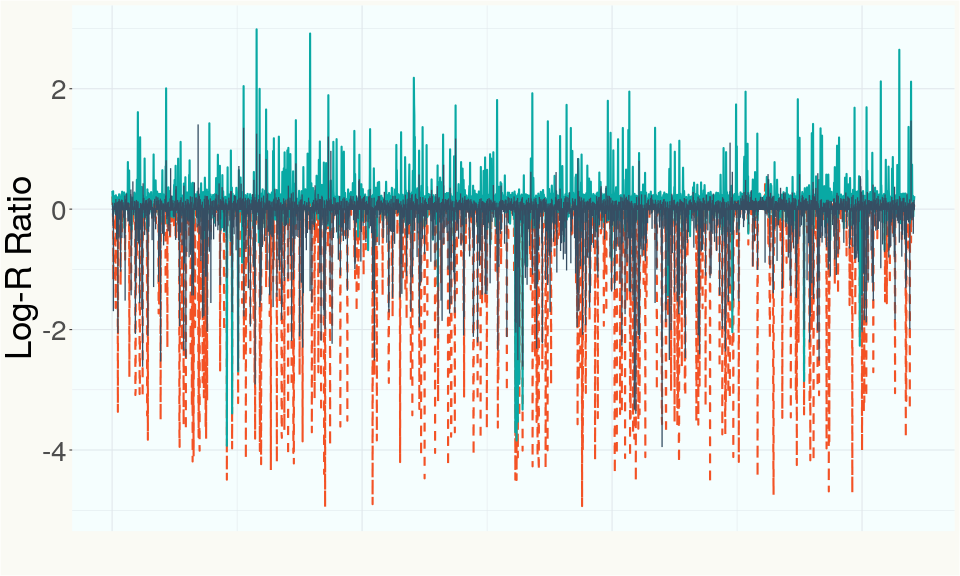
Summary of Deletions
THCAS
- Correlation:
- 0.97
- Call:
- deleted
CBDAS
- Correlation:
- 0.27
- Call:
- intact
CBCAS
- Correlation:
- -0.08
- Call:
- intact
Plant Sex
- Correlation:
- 0.73
- Call:
- female
Variants (THCAS, CBDAS, and CBCAS)
| CBDAS | c.221C>G | p.Thr74Ser | missense variant | moderate | contig1772 | 2082447 | C/G | |
| CBDAS | c.407G>A | p.Arg136His | missense variant | moderate | contig1772 | 2082633 | G/A | |
| CBDAS | c.1420A>C | p.Lys474Gln | missense variant | moderate | contig1772 | 2083646 | A/C | |
| CBDAS | c.1628G>A | p.Arg543His | missense variant | moderate | contig1772 | 2083854 | G/A |
|
Variants (Select Genes of Interest)
| GPPs1 | c.744C>G | p.Asp248Glu | missense variant | moderate | contig676 | 168993 | C/G |
|
| EMF1-2 | c.710A>C | p.His237Pro | missense variant | moderate | contig885 | 810 | A/C | |
| PHL-2 | c.44G>A | p.Arg15Lys | missense variant | moderate | contig2621 | 337613 | G/A | |
| PHL-2 | c.932T>C | p.Leu311Pro | missense variant | moderate | contig2621 | 340210 | T/C | |
| PHL-2 | c.1096G>A | p.Ala366Thr | missense variant | moderate | contig2621 | 340374 | G/A | |
| PHL-2 | c.2564T>A | p.Phe855Tyr | missense variant | moderate | contig2621 | 342607 | T/A | |
| PHL-2 | c.2624C>T | p.Ser875Phe | missense variant | moderate | contig2621 | 342667 | C/T | |
| PHL-2 | c.2756A>C | p.Glu919Ala | missense variant | moderate | contig2621 | 342799 | A/C | |
| PHL-2 | c.2933G>T | p.Arg978Leu | missense variant | moderate | contig2621 | 342976 | G/T | |
| PHL-2 | c.3002A>G | p.Tyr1001Cys | missense variant | moderate | contig2621 | 343045 | A/G | |
| PHL-2 | c.3027G>T | p.Lys1009Asn | missense variant | moderate | contig2621 | 343070 | G/T | |
| PHL-2 | c.3033T>G | p.Cys1011Trp | missense variant | moderate | contig2621 | 343076 | T/G | |
| PHL-2 | c.3209A>G | p.Gln1070Arg | missense variant | moderate | contig2621 | 343252 | A/G | |
| PKSG-4a | c.1000T>C | p.Tyr334His | missense variant | moderate | contig700 | 1938411 | T/C | |
| PKSG-2a | c.1152T>A | p.Asn384Lys | missense variant | moderate | contig700 | 1944238 | A/T | |
| PKSG-2a | c.224A>G | p.Lys75Arg | missense variant | moderate | contig700 | 1945166 | T/C | |
| PKSG-2b | c.774G>A | p.Met258Ile | missense variant | moderate | contig700 | 1950864 | C/T | |
| PKSG-2b | c.230T>C | p.Val77Ala | missense variant | moderate | contig700 | 1951408 | A/G | |
| PKSG-2b | c.224A>G | p.Lys75Arg | missense variant | moderate | contig700 | 1951414 | T/C | |
| PKSG-2b | c.167C>G | p.Thr56Ser | missense variant | moderate | contig700 | 1951471 | G/C | |
| PKSG-2b | c.31A>T | p.Thr11Ser | missense variant | moderate | contig700 | 1951851 | T/A | |
| PKSG-4b | c.496A>G | p.Lys166Glu | missense variant | moderate | contig700 | 2721177 | T/C | |
| PKSG-4b | c.431T>G | p.Val144Gly | missense variant | moderate | contig700 | 2721242 | A/C | |
| PKSG-4b | c.419A>G | p.Asp140Gly | missense variant | moderate | contig700 | 2721254 | T/C | |
| PKSG-4b | c.316+2T>A | splice donor variant & intron variant | high | contig700 | 2723818 | A/T | ||
| DXR-2 | c.431C>G | p.Ala144Gly | missense variant | moderate | contig380 | 287760 | G/C | |
| ELF3 | c.812G>C | p.Gly271Ala | missense variant | moderate | contig97 | 242518 | G/C | |
| ELF3 | c.1366T>G | p.Leu456Val | missense variant | moderate | contig97 | 244197 | T/G |
|
| ELF3 | c.1630A>G | p.Thr544Ala | missense variant | moderate | contig97 | 244461 | A/G | |
| ELF3 | c.2198G>T | p.Arg733Leu | missense variant | moderate | contig97 | 245029 | G/T | |
| HDS-2 | c.679G>C | p.Gly227Arg | missense variant | moderate | contig95 | 1990632 | G/C |
|
| PHL-1 | c.2551A>G | p.Thr851Ala | missense variant | moderate | contig1439 | 1487246 | T/C | |
| PHL-1 | c.1387A>G | p.Thr463Ala | missense variant | moderate | contig1439 | 1489811 | T/C | |
| TFL1 | c.302-1G>A | splice acceptor variant & intron variant | high | contig1636 | 520616 | C/T | ||
| HDS-1 | c.1618A>G | p.Ile540Val | missense variant | moderate | contig1891 | 885936 | T/C | |
| HDS-1 | c.136G>A | p.Val46Ile | missense variant | moderate | contig1891 | 889256 | C/T | |
| HDS-1 | c.35G>A | p.Cys12Tyr | missense variant | moderate | contig1891 | 889357 | C/T | |
| PIE1-2 | c.5932A>G | p.Ile1978Val | missense variant | moderate | contig1460 | 1185552 | T/C | |
| PIE1-2 | c.5884G>A | p.Gly1962Ser | missense variant | moderate | contig1460 | 1185715 | C/T | |
| PIE1-2 | c.4721T>C | p.Val1574Ala | missense variant | moderate | contig1460 | 1187319 | A/G |
|
| PIE1-2 | c.3554G>A | p.Arg1185Lys | missense variant | moderate | contig1460 | 1188486 | C/T | |
| PIE1-2 | c.3506A>C | p.Glu1169Ala | missense variant | moderate | contig1460 | 1188534 | T/G | |
| PIE1-2 | c.2188G>A | p.Ala730Thr | missense variant | moderate | contig1460 | 1189852 | C/T | |
| PIE1-2 |
c.2083_2085d |
p.Val695del | conservative inframe deletion | moderate | contig1460 | 1189954 | GGAC/G | |
| PIE1-2 | c.2072A>G | p.His691Arg | missense variant | moderate | contig1460 | 1189968 | T/C | |
| PIE1-2 | c.2027A>T | p.Gln676Leu | missense variant | moderate | contig1460 | 1190013 | T/A | |
| PIE1-2 | c.1872T>A | p.Asp624Glu | missense variant | moderate | contig1460 | 1190252 | A/T | |
| PIE1-2 | c.1117C>G | p.Gln373Glu | missense variant | moderate | contig1460 | 1192281 | G/C | |
| PIE1-2 | c.1093G>A | p.Gly365Ser | missense variant | moderate | contig1460 | 1192305 | C/T | |
| PIE1-2 | c.710C>T | p.Pro237Leu | missense variant | moderate | contig1460 | 1193804 | G/A | |
| PIE1-2 | c.706T>C | p.Tyr236His | missense variant | moderate | contig1460 | 1193808 | A/G | |
| PIE1-2 | c.637T>A | p.Ser213Thr | missense variant | moderate | contig1460 | 1194421 | A/T | |
| PIE1-2 | c.349C>T | p.Pro117Ser | missense variant & splice region variant | moderate | contig1460 | 1195017 | G/A |
|
| EMF2 | c.1772A>G | p.Gln591Arg | missense variant | moderate | contig954 | 3059929 | A/G | |
| EMF1-1 | c.242A>G | p.Lys81Arg | missense variant | moderate | contig883 | 269731 | A/G | |
| FLD | c.2681T>C | p.Ile894Thr | missense variant | moderate | contig1450 | 2044853 | A/G | |
| PIE1-1 | c.742T>A | p.Ser248Thr | missense variant | moderate | contig1225 | 2279320 | T/A | |
| PIE1-1 | c.773A>G | p.Asn258Ser | missense variant & splice region variant | moderate | contig1225 | 2279897 | A/G | |
| PIE1-1 | c.811T>C | p.Tyr271His | missense variant | moderate | contig1225 | 2279935 | T/C | |
| PIE1-1 | c.815C>T | p.Pro272Leu | missense variant | moderate | contig1225 | 2279939 | C/T | |
| PIE1-1 | c.1222C>G | p.Gln408Glu | missense variant | moderate | contig1225 | 2281482 | C/G | |
| PIE1-1 | c.1394A>G | p.Asp465Gly | missense variant | moderate | contig1225 | 2281654 | A/G | |
| PIE1-1 | c.1454T>C | p.Val485Ala | missense variant | moderate | contig1225 | 2281714 | T/C |
|
| PIE1-1 |
c.1548_1549i |
p.Gln516_Glu |
conservative inframe insertion | moderate | contig1225 | 2281807 | A/AGAT | |
| PIE1-1 | c.2174A>G | p.His725Arg | missense variant | moderate | contig1225 | 2283789 | A/G |
|
| PIE1-1 |
c.2185_2187d |
p.Val729del | conservative inframe deletion | moderate | contig1225 | 2283796 | AGTC/A |
|
| PIE1-1 | c.3608A>C | p.Glu1203Ala | missense variant | moderate | contig1225 | 2285223 | A/C | |
| PIE1-1 | c.3656G>A | p.Arg1219Lys | missense variant | moderate | contig1225 | 2285271 | G/A | |
| PIE1-1 | c.3869A>C | p.Asn1290Thr | missense variant | moderate | contig1225 | 2285484 | A/C | |
| PIE1-1 | c.4823T>C | p.Val1608Ala | missense variant | moderate | contig1225 | 2286438 | T/C |
|
| PIE1-1 | c.6041T>C | p.Met2014Thr | missense variant | moderate | contig1225 | 2288214 | T/C |
|
| PIE1-1 | c.6738G>T | p.Glu2246Asp | missense variant | moderate | contig1225 | 2289303 | G/T |
|
| GGR | c.317C>T | p.Pro106Leu | missense variant | moderate | contig2282 | 549309 | C/T | |
| GGR | c.376G>C | p.Glu126Gln | missense variant | moderate | contig2282 | 549368 | G/C | |
| GGR | c.541G>A | p.Val181Ile | missense variant | moderate | contig2282 | 549533 | G/A |
|
Nearest genetic relatives (All Samples)
- 0.111 BlueBerry Cheesecake x JL Male (RSP11201)
- 0.115 Fedora 17 (RSP11203)
- 0.118 Carmagnola USO 31 (RSP11204)
- 0.121 Carmaleonte (RSP11207)
- 0.127 Tiborszallasie (RSP11210)
- 0.144 Eletta Campana (RSP11209)
- 0.152 CS (RSP11208)
- 0.152 Feral (RSP11206)
- 0.154 Carmagnola garden (RSP11603)
- 0.155 Electra (RSP11366)
- 0.157 Herijuana (RSP11181)
- 0.160 JL Cross 78 (RSP11579)
- 0.167 Thank You Jerry (RSP11459)
- 0.172 JL Cross 80 (RSP11581)
- 0.173 JL Cross 77 (RSP11578)
- 0.174 Wedding Cake x MAC (RSP11464)
- 0.174 AVIDEKEL USA (RSP11169)
- 0.175 Lift (RSP11378)
- 0.178 Master Kush (RSP11182)
- 0.178 JL Cross 34 (RSP11535)
Most genetically distant strains (All Samples)
- 0.233 80E (RSP11212)
- 0.231 80E (RSP11213)
- 0.227 80E (RSP11211)
- 0.219 JL 3rd Gen Father (RSP11196)
- 0.216 TI-7 (RSP11601)
- 0.216 Cbot-2019-005 (RSP11133)
- 0.215 Chem 91 (RSP11185)
- 0.215 JL 4th Gen 6 (RSP11200)
- 0.213 UnObtanium (RSP11611)
- 0.212 JL 4th Gen 2 (RSP11194)
- 0.211 Unknown- Cherry Wine - 001 (RSP11268)
- 0.210 Chematonic Cannatonic x Chemdawg (RSP11394)
- 0.209 Fatso (RSP11740)
- 0.209 JL 4th Gen 1 (RSP11193)
- 0.208 Headcheese (RSP11192)
- 0.207 Red Eye OG (RSP11190)
- 0.207 Legend OG TC (RSP12584)
- 0.206 unknown (RSP11432)
- 0.206 Pineapple Haze (RSP11652)
- 0.206 Unknown- Cherry Wine - 003 (RSP11270)
Nearest genetic relative in Phylos dataset
- Overlapping SNPs:
- 54
- Concordance:
- 41
Nearest genetic relative in Lynch dataset
- Overlapping SNPs:
- 1
- Concordance:
- 1
Blockchain Registration Information
- Transaction ID
-
a9429a8783c83efe
fcdaff34881a9b9a 4470d66305647879 a5ffa3843aa2e023 - Stamping Certificate
- Download PDF (39.6 KB)
- SHASUM Hash
-
bd99880009b7c2abae2b39bddfec13f6 a347938d35f611e2 157c8c6089731d2a