JL Cross 33
RSP 11534
Grower: Kevin McKernan
General Information
- Accession Date
- June 21, 2020
- Reported Plant Sex
- not reported
- Report Type
- CannSNP90
The strain rarity visualization shows how distant the strain is from the other cultivars in the Kannapedia database. The y-axis represents genetic distance, getting farther as you go up. The width of the visualization at any position along the y-axis shows how many strains there are in the database at that genetic distance. So, a common strain will have a more bottom-heavy shape, while uncommon and rare cultivars will have a visualization that is generally shifted towards the top.
Chemical Information
Cannabinoid and terpenoid information provided by the grower.
Cannabinoids
No information provided.
Terpenoids
No information provided.
Genetic Information
- Plant Type
- Type II
File Downloads
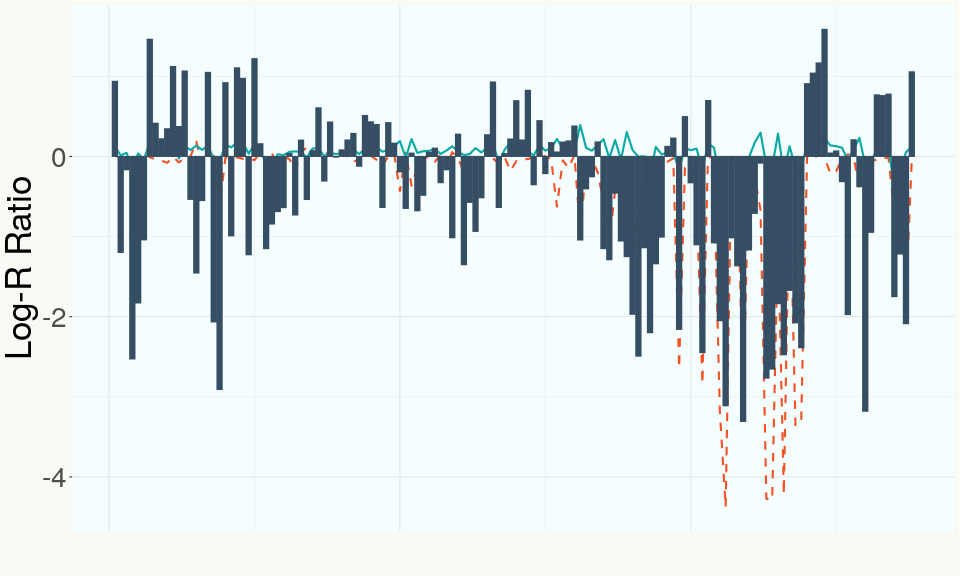
This chart represents the Log-R Ratio (LRR) over variants in the region of the THCA synthase gene. A high correlation between the LRR and samples with a known deletion in THCA synthase indicates the THCA region is deleted and a low correlation indicates it is intact.
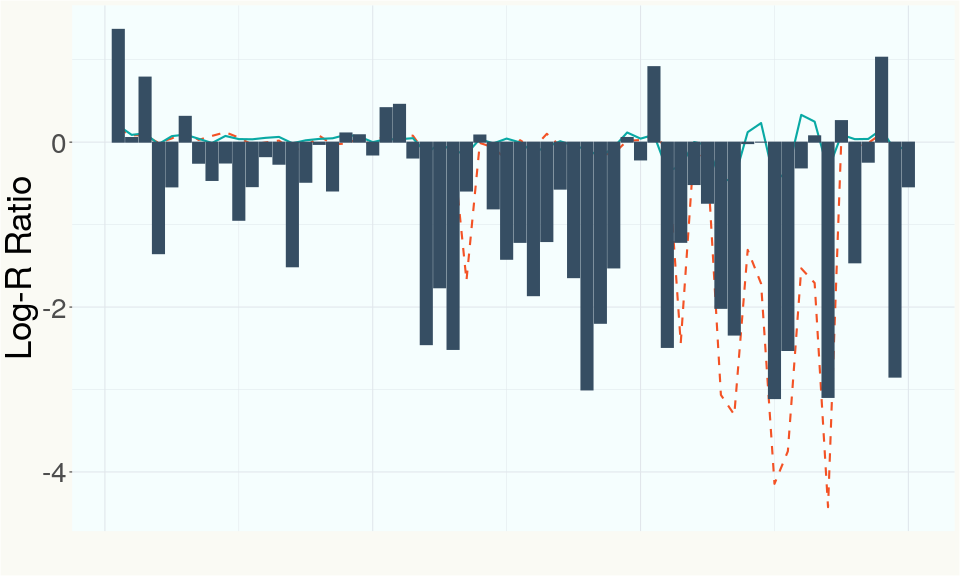
This chart represents the Log-R Ratio (LRR) over variants in the region of the CBDA synthase gene. A high correlation between the LRR and samples with a known deletion in CBDA synthase indicates the CBDA region is deleted and a low correlation indicates it is intact.
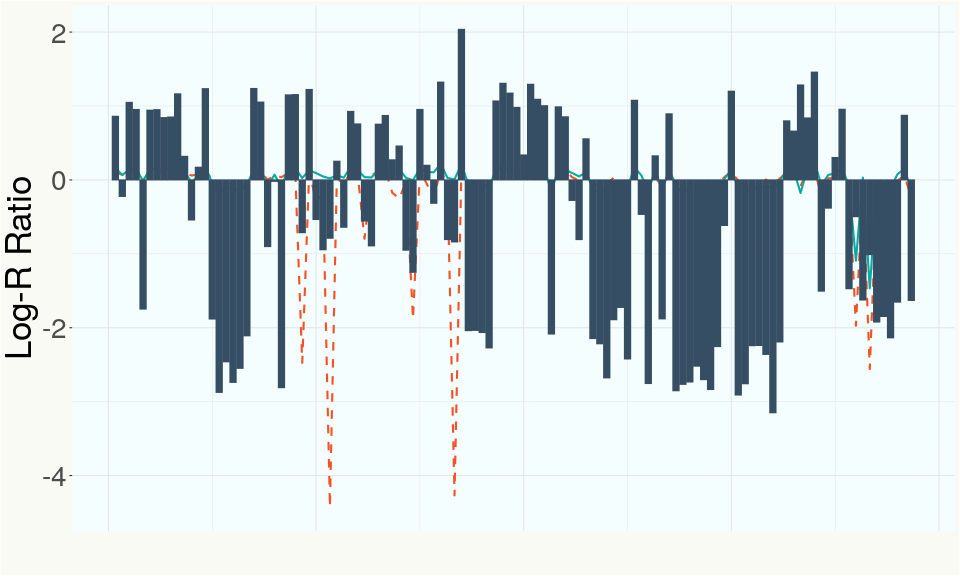
This chart represents the Log-R Ratio (LRR) over variants in the region of the CBCA synthase gene. A high correlation between the LRR and samples with a known deletion in CBCA synthase indicates the CBCA region is deleted and a low correlation indicates it is intact.
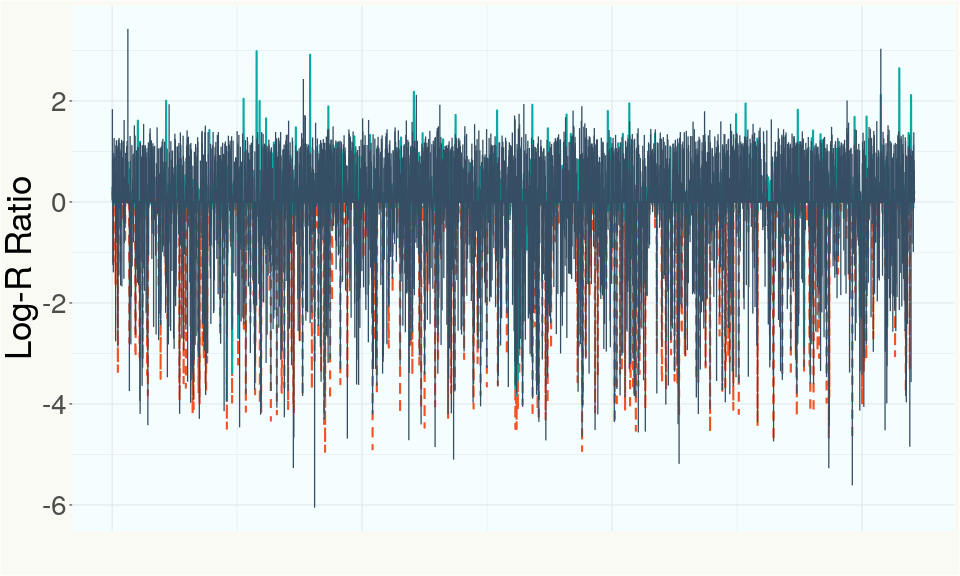
This chart represents the Log-R Ratio (LRR) over variants in the Y-contigs. A high correlation between the LRR and samples which are known Females indicates these Y-contigs are deleted in this sample and a low correlation indicates that the Y-contigs are not deleted and is likely Male.
Summary of Deletions
THCAS
- Correlation:
- 0.62
- Call:
- intact
CBDAS
- Correlation:
- 0.47
- Call:
- intact
CBCAS
- Correlation:
- 0.14
- Call:
- intact
Plant Sex
- Correlation:
- 0.7
- Call:
- female
Variants (THCAS, CBDAS, and CBCAS)
| CBDAS | c.407G>A | p.Arg136His | missense variant | moderate | contig1772 | 2082633 | G/A | |
| CBDAS | c.503A>G | p.Asn168Ser | missense variant | moderate | contig1772 | 2082729 | A/G | |
| CBDAS | c.688T>A | p.Leu230Ile | missense variant | moderate | contig1772 | 2082914 | T/A |
|
| CBDAS | c.704C>G | p.Ala235Gly | missense variant | moderate | contig1772 | 2082930 | C/G |
|
| THCAS | c.355A>T | p.Met119Leu | missense variant | moderate | contig741 | 4417473 | T/A |
|
Variants (Select Genes of Interest)
| PHL-2 | c.3002A>G | p.Tyr1001Cys | missense variant | moderate | contig2621 | 343045 | A/G | |
| PKSG-2a | c.67T>A | p.Phe23Ile | missense variant | moderate | contig700 | 1945567 | A/T | |
| PKSG-2a | c.31A>T | p.Thr11Ser | missense variant | moderate | contig700 | 1945603 | T/A | |
| PKSG-2b | c.31A>T | p.Thr11Ser | missense variant | moderate | contig700 | 1951851 | T/A | |
| aPT1 | c.406A>G | p.Ile136Val | missense variant | moderate | contig121 | 2839605 | A/G | |
| aPT1 | c.629C>T | p.Thr210Ile | missense variant | moderate | contig121 | 2840237 | C/T | |
| aPT1 | c.727G>T | p.Glu243* | stop gained | high | contig121 | 2841362 | G/T | |
| aPT1 | c.864C>G | p.Asn288Lys | missense variant | moderate | contig121 | 2842407 | C/G | |
| HDS-1 | c.35G>A | p.Cys12Tyr | missense variant | moderate | contig1891 | 889357 | C/T | |
| PIE1-2 | c.2188G>A | p.Ala730Thr | missense variant | moderate | contig1460 | 1189852 | C/T | |
| PIE1-2 | c.1872T>A | p.Asp624Glu | missense variant | moderate | contig1460 | 1190252 | A/T | |
| PIE1-2 | c.982G>A | p.Glu328Lys | missense variant | moderate | contig1460 | 1192416 | C/T | |
| PIE1-2 | c.710C>T | p.Pro237Leu | missense variant | moderate | contig1460 | 1193804 | G/A | |
| PIE1-2 | c.706T>C | p.Tyr236His | missense variant | moderate | contig1460 | 1193808 | A/G | |
| EMF2 | c.722C>T | p.Thr241Ile | missense variant | moderate | contig954 | 3050302 | C/T | |
| FT | c.196A>G | p.Ile66Val | missense variant | moderate | contig1561 | 3124620 | A/G | |
| FT | c.419G>A | p.Ser140Asn | missense variant | moderate | contig1561 | 3126658 | G/A | |
| FT | c.440A>C |
p.Ter147Sere |
stop lost & splice region variant | high | contig1561 | 3126679 | A/C | |
| FLD | c.125G>A | p.Ser42Asn | missense variant | moderate | contig1450 | 2047909 | C/T | |
| PIE1-1 | c.815C>T | p.Pro272Leu | missense variant | moderate | contig1225 | 2279939 | C/T | |
| PIE1-1 | c.1454T>C | p.Val485Ala | missense variant | moderate | contig1225 | 2281714 | T/C |
|
Nearest genetic relatives (All Samples)
- 0.046 JL Cross 26 (RSP11527)
- 0.064 JL Cross 34 (RSP11535)
- 0.065 Pineapple Haze (RSP11652)
- 0.077 JL Cross 24 (RSP11525)
- 0.083 JL Cross 15 (RSP11516)
- 0.085 JL Cross 27 (RSP11528)
- 0.096 JL Cross 17 (RSP11518)
- 0.096 JL Cross 25 (RSP11526)
- 0.101 JL Cross 31 (RSP11532)
- 0.111 JL Cross 28 (RSP11529)
- 0.111 JL Cross 9 (RSP11510)
- 0.111 JL Cross 18 (RSP11519)
- 0.112 JL X NSPM1 14 (RSP11473)
- 0.113 TI-9 (RSP11609)
- 0.115 JL Cross 4 (RSP11505)
- 0.116 JL Cross 80 (RSP11581)
- 0.130 JL Cross 32 (RSP11533)
- 0.139 JL X NSPM1 5 (RSP11467)
- 0.139 JL Cross 21 (RSP11522)
- 0.140 JL Cross 29 (RSP11530)
Most genetically distant strains (All Samples)
- 0.243 80E (RSP11212)
- 0.240 80E (RSP11211)
- 0.240 CS (RSP11208)
- 0.239 80E (RSP11213)
- 0.236 Tiborszallasie (RSP11210)
- 0.235 Fedora 17 (RSP11203)
- 0.234 Feral (RSP11205)
- 0.230 Carmaleonte (RSP11207)
- 0.228 Eletta Campana (RSP11209)
- 0.227 Arcata Trainwreck (RSP11176)
- 0.224 Carmagnola USO 31 (RSP11204)
- 0.224 Unknown- Cherry Wine - 001 (RSP11268)
- 0.223 Feral (RSP11206)
- 0.222 Cbot-2019-005 (RSP11133)
- 0.221 Unknown- Cherry Wine - 002 (RSP11269)
- 0.220 Unknown- Cherry Wine - 003 (RSP11270)
- 0.220 Goomendaze (RSP11462)
- 0.219 Swiss Dream (RSP11408)
- 0.219 AVIDEKEL 2 0 (RSP11174)
- 0.218 Unknown- Cherry Wine - 004 (RSP11271)
Nearest genetic relative in Phylos dataset
- Overlapping SNPs:
- 17
- Concordance:
- 12
Nearest genetic relative in Lynch dataset
- Overlapping SNPs:
- 5
- Concordance:
- 4
Blockchain Registration Information
- SHASUM Hash
-
2f3595096bbde764264b18b77fe3bae3 13d15b5aa33bd6ae 244925a093b5782f